Submitted by M. Rodrigues on Thu, 06/05/2021 - 15:17
Threshold accumulation of a constitutive protein explains E. coli cell-division behavior in nutrient upshifts
The work recently published in Proc. Natl. Acad. Sci. (USA) explores the dynamic adaptation of cell-division control in bacteria to shifts in the growth conditions.
The cell cycle is one of those questions that is always at the forefront of biology, and today the high throughput imaging of large number of cell cycles resolved at the single cell level makes new discoveries possible. Despite a boost of recent progress in dynamic single-cell measurements and analyses in Escherichia coli, we still lack a mechanistic understanding of the determinants of the cell's decision to divide.
Specifically, the debate is open regarding the processes linking growth and chromosome replication to division and on the molecular origin of the observed “adder correlations,” whereby cells divide after having added a roughly constant volume, independent of their initial volume.
In order to gain insight into these questions, the experiments of Mia Panlilio have interrogated dynamic size-growth behaviour of single cells across nutrient upshifts with a high-precision microfluidic device. The team find that the division rate reacts quickly after nutrients change, much before the growth rate goes to a steady state, and in such a way that adder correlations are robustly conserved throughout the transition.
Pietro Cicuta, co-last author, explains that: “This publication is a significant part of Mia Panlilio’s PhD work. Mia worked as an extremely talented biological physicist, under what turned out to be really challenging years for her – we’re all particularly proud of the data and insights she was able to produce”. The work also represents the long term fruits of two back-to-back HFSP grants on which Pietro, theoretical physicist Marco Cosentino Lagomarsino (now at University of Milan, and IFOM, Italy) and microbiologist Bianca Sclavi (CNRS, Paris) collaborated to address a number of biological questions in E.coli with physics tools and concepts.
Bianca Sclavi adds: “The mechanism leading to cell division in the bacterium Escherichia coli is unknown, but we know that it results in adding a roughly constant size every cell cycle, regardless of size at birth. While most available studies try to infer information on cell division from steadily dividing cells in constant nutrient conditions, this study leverages a high-resolution device to monitor single-cell growth division upon nutrient changes.”
The work includes a biological physics modelling component, led by Marco Cosentino Lagomarsino, who says: “The comparison of these non-equilibrium, transient, data to simple mathematical models allowed us to falsify proposed mechanisms, where replication–segregation or septum completions are the limiting step for cell division. Instead, we showed that the accumulation of a putative constitutively expressed “P-sector divisor” protein to a threshold would explain the behaviour during the shift.”
This work brings us a step closer to understanding the fundamental links of how two of the basic “modules” of life in a cell, gene expression and growth, regulate cell division.
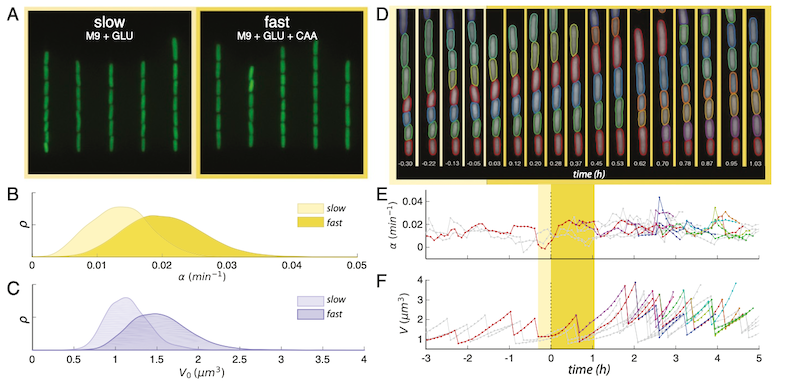
FIG. 1. Central to the work is the robust and long term single cell tracking through a nutritional upshift. A. Snapshots of trapped cells in microfluidic device under stringent (left) and rich nutrient conditions (right) (false-coloured). B. Steady state distributions of growth rate α in the two growth media. C. Steadystate distributions of birth volumes V0 for the sample populations in B. D. Sample segmentation and tracking before and after the growth media switch is implemented, time since switch shown in hours. E. Tracking the change of instantaneous growth rate following the switch for the cells shown above. F. Volume tracking of the same cell lines.
Reference: Panlilio, M., et al. “Threshold accumulation of a constitutive protein explains E. coli cell-division behavior in nutrient upshifts”. Proc. Natl. Acad. Sci. (USA) 118, No 18 e2016391118 (2021). https://doi.org/10.1073/pnas.2016391118