Submitted by M. Rodrigues on Wed, 29/09/2021 - 18:26
Liquid–liquid phase separation underpins the formation of replication factories in rotaviruses
Dr Alexander Borodavka (Dept Biochemistry) coordinated a multidisciplinary project on the mechanism of formation of viral replication factories, also known as viroplasms. Using advanced microscopy techniques including DNA-PAINT, high-throughput analyses of protein condensates and viral reverse genetics approaches the authors reveal for the first time how two nonstructural proteins, form liquid–liquid phase-separated condensates as the structural foundation of viroplasms and highlight the importance of exploiting molecular condensates as antiviral therapeutic targets. Work published in EMBO J.
Many RNA viruses induce the formation of subcellular organelles that support their replication, also known as viral replication factories, or viroplasms. “Rotaviruses, famous for causing gastroenteritis in children, are remarkably complex viruses containing segmented RNA genomes of eleven double-stranded RNA molecules that serve as viral chromosomes. The precise molecular mechanism behind the assembly of rotaviruses remains a mystery”, explains Dr Borodavka senior author of the paper.
The two important proteins studied in this work are:
- NSP2: RNA chaperone, believed to be the key protein that governs sequence-specific RNA assembly of viral transcripts-precursors and accumulates within the viroplasms.
- NSP5: non-structural phosphoprotein also present in viroplasms.
Both NSP2 and NSP5 are not packaged into mature virions and thus are termed ‘nonstructural’.
“How viroplasms are formed, and their dynamic role in supporting rotavirus replication remains poorly understood”, says Dr Borodavka.
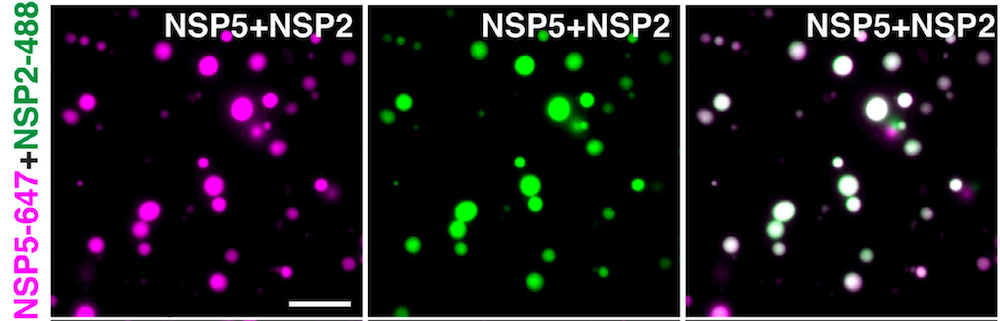
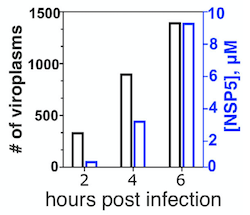
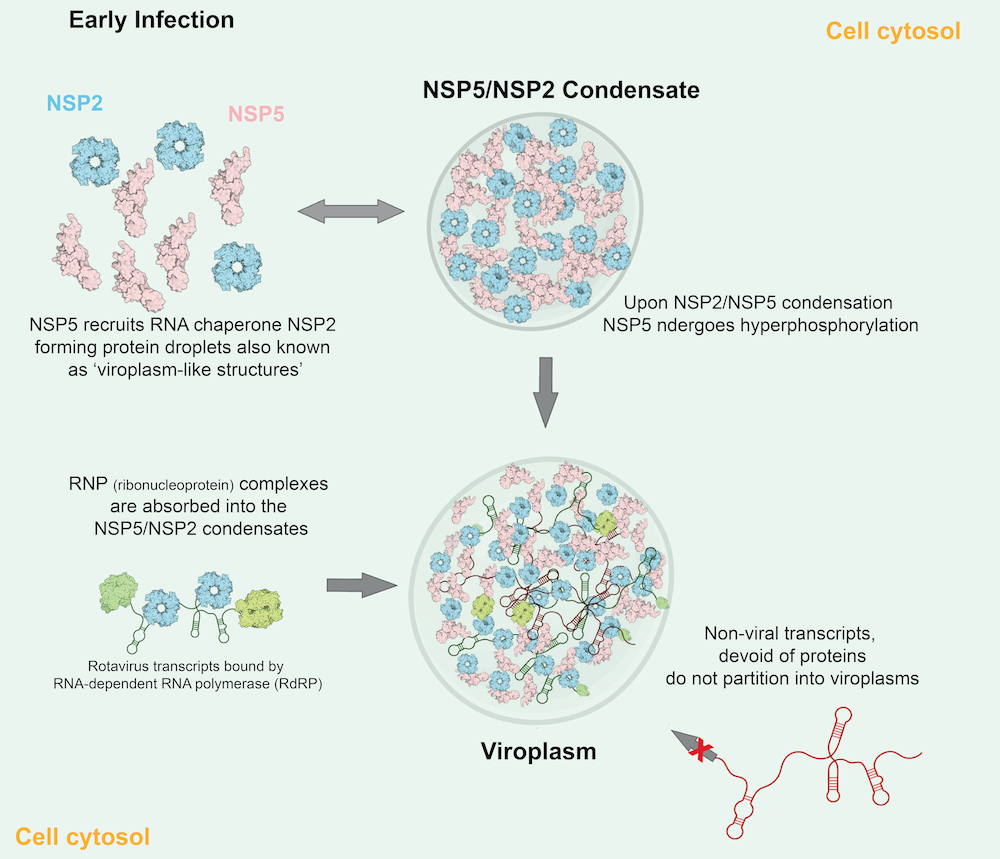
Julia Acker, co-first author tells, “- We discovered that membraneless replication factories of rotaviruses represent complex protein-RNA condensates that are initially formed via liquid–liquid phase separation of the viroplasm-forming proteins NSP5 and rotavirus RNA chaperone NSP2.” The authors used live imaging of rotavirus-infected cells and microscopic analysis of recombinantly expressed fluorescently labelled rotavirus non-structural proteins, and demonstrated that NSP2/NSP5 form condensates at physiologically relevant low micromolar concentrations achieved in the cytoplasm of virus-infected cells.
Early infection stage condensates could be reversibly dissolved by 1,6-hexanediol, and, remarkably, by far less toxic propylene glycol, that released rotavirus transcripts from these condensates. Interestingly, during the early stages of infection, propylene glycol treatments reduced viral replication and phosphorylation of the condensate-forming protein NSP5. However, during late infection, viral condensates exhibited altered material properties and became resistant to propylene glycol, coinciding with hyperphosphorylation of NSP5.
Collectively, data suggests that rotavirus replicative condensate formation requires NSP5 as a scaffold protein that undergoes phase-separation in the presence of the RNA chaperone NSP2. Oligomerisation of NSP5 is crucial for its phase-separation, albeit non-oligomerising mutants of NSP5 retain their ability to partition into pre-formed NSP5/NSP2 condensates.
Implications
“Phase transitions of viral proteins have been also recently described for SARS-CoV2, respiratory syncytial, measles and rabies viruses to name a few. It is clear that ccompounds that disrupt phase-separation of viral replicative condensates by shifting the phase boundary of condensates, or by promoting hardening of condensates, can be exploited for future antiviral strategies”, explains Dr Borodavka.
The viral RNA/protein condensates involved in replication of viral RNA genomes represent an attractive target for developing new antiviral interventions.
“These findings set the foundation for future studies to improve our understanding of the role of phase transitions in viral replication in different viruses and hosts. Such studies will hugely benefit from recent advances in quantitative microscopy, high-throughput analyses of individual condensate droplets and viral reverse genetics approaches.”, highlights Dr Borodavka.
Proposed model of LLPS-driven formation of viral replication factories in rotaviruses
This research was funded by Wellcome Trust.