Alexander Ohmann
Outperforming nature with a lipid-flipping enzyme built from DNA
Keywords: DNA nanotechnology, membrane transport, DNA origami, confocal microscopy, lipid scrambling
Membrane proteins are key for every cell’s health. Neuronal diseases like Alzheimer’s and blood disorders like sickle cell disease have been linked to impaired membrane protein function. About 30 % of the human genome encodes membrane proteins and roughly 50 % of approved drugs are targeted at them. What if we could give a patient something that can take over the role of the defective protein? Programmable medicine that could mimic the protein’s function? This vision drove the research of my PhD.
In healthy cells, the lipid composition of cell membranes is typically asymmetric, meaning that there are different types of lipids in the outer and in the inner bilayer leaflet. However, certain events use lipids as signals to the extracellular environment, for example for inducing controlled death (apoptosis) in cells that do not function properly. Membrane proteins termed ‘lipid scramblases’ are then activated and rapidly ‘flip lipids’ between the bilayer leaflets to equilibrate the lipid composition.
To create a synthetic, controllable counterpart, we employed DNA nanotechnology. DNA is hereby not used for storing genetic information but as a building material. Using a method called ‘DNA origami’, billions of nanometer-sized structures with precisely controlled geometries can assemble themselves in a test tube. Chemically modifying them with cholesterol then facilitates their interaction with lipid membranes. Using this approach, I designed a DNA-built enzyme that can self-insert into lipid bilayers and induce rapid lipid mixing similar to natural scramblases.
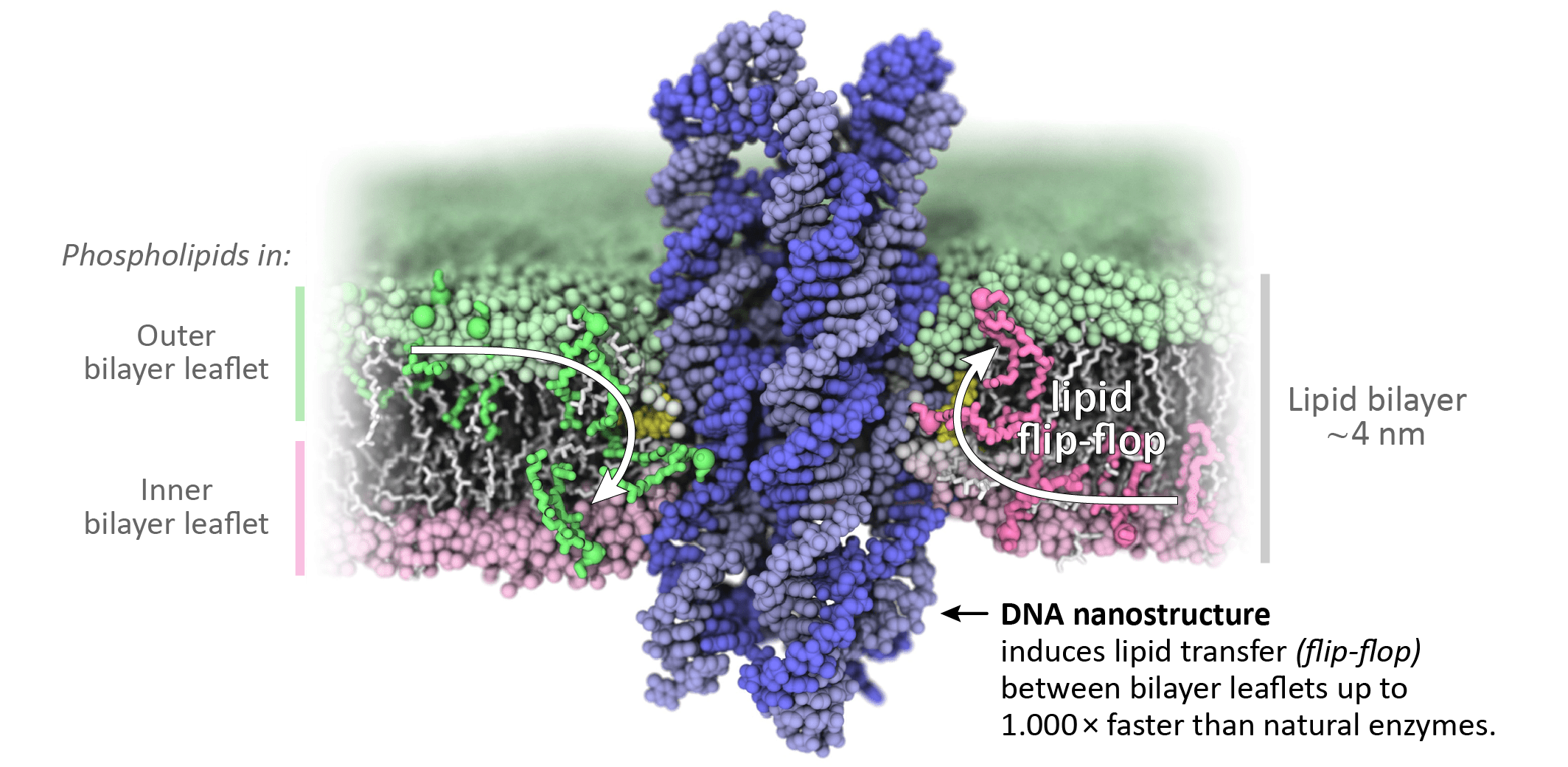
To understand the interaction of our DNA structures (blue in image) with the lipids in the bilayer leaflets (pink and green), we collaborated with Aleksei Aksimentiev’s group from the University of Illinois Urbana-Champaign. Their all-atom molecular dynamics simulations revealed that the membrane-inserted DNA structures cause the lipid head groups to reorient and form a toroidal lipid pore at the DNA-lipid interface. This toroidal pore creates a connection between the bilayer leaflets allowing lipids to rapidly transfer between them (= flip-flop). Astonishingly, this happens at a rate of 10,000,000 lipids per second, about 1,000-fold faster than reported rates for natural scramblases.
In the Keyser group, we then confirmed these simulations experimentally. We created artificial lipid vesicles to mimic cell membranes and labelled some of the lipids with a fluorescent marker. Using fluorescence microscopy, we then determined the DNA-induced lipid flipping rates between the bilayer leaflets. Strikingly, our results similarly yielded flipping rates of 107 lipids per second. We then applied our DNA scramblases to human breast cancer cells and observed indications that our constructs can indeed induce lipid flipping and apoptosis even in biological cell membranes.
While there is still a lot more to learn, pioneering the use of DNA nanostructures for controlling the lipid composition of biological membranes opens new avenues for innovative applications in biophysical research and medicine.
Alexander's Google Scholar
Keyser Lab - Department of Physics