Submitted by M. Rodrigues on Mon, 05/07/2021 - 10:58
Virus-host interactions shape viral dispersal giving rise to distinct classes of traveling waves in spatial expansions
The work recently published by the group of Dr Diana Fusco (Dept Physics) in Physical Review X explores the genetic diversity of viral expansions using experiments with bacteriophage and simulations.
Spatial range expansions, where populations grow and spread to new territory, are a ubiquitous biological process, from invasive plant species to ancient human migration. Typically, these expansions can be classed as either ‘pulled,’ where the expansion is led by individuals at the perimeter of the population, or ‘pushed,’ where cooperation between individuals drives the expansion more from within.
Michael Hunter, first author of the study, explains that: “Using a model ecosystem of phage T7 and its bacterial host E. coli, we show with a combination of experiments and simulations for the first time, that viral expansions can transition from pulled to pushed, even though no cooperation between viral particles exists.” Authors observed that the transition spontaneously emerges from the interplay between viral dispersal and infection dynamic: viral dispersal depends on the host density, which, in turn, is continuously modified by the virus as the expansion advances. With these results the team uncovered two independent physical mechanisms that contribute to this interplay: one is linked to the steric interactions between virus and host in crowded environments; the second is linked to the viral incubation time, during which the virus is trapped inside the host unable to diffuse.
"Our results point to the phage-bacteria system as a highly controllable platform to experimentally investigate the dynamics of expanding populations with density-dependent dispersal, with applications both in eco-evolutionary dynamics and in the study of pattern formation", says Diana Fusco, last author of the paper. Also, beyond phages, our findings suggest that expanding viral populations retain genetic diversity much longer than previously thought, suggesting that they can easily adapt to changes in the environment.
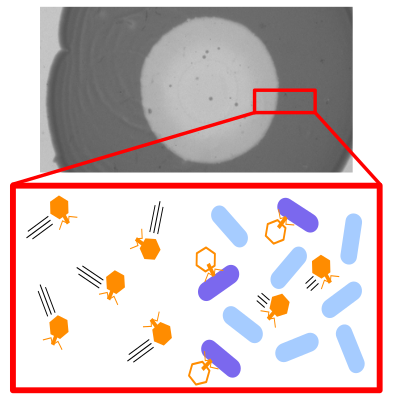
Graphical abstract: As the viral expansion proceeds, infected cells lyse (plaque formation, top), which inevitably generates a non-uniform bacterial density profile where susceptible host is depleted in the bulk of the viral expansion (red triangle, left side). This heterogeneity in host density facilitates viral dispersal in the bulk, as cells hinder viral diffusion, allowing the viruses in the back of the expansion to catch up with those at the front (pushed wave.
Video: Bacterial colony (left) cleared by phage. Total time 24h.
Reference: Michael Hunter, Nikhil Krishnan, Tongfei Liu, Wolfram Möbius, and Diana Fusco. “Virus-Host Interactions Shape Viral Dispersal Giving Rise to Distinct Classes of Traveling Waves in Spatial Expansions”. Phys. Rev. X 11, 021066 (2021). https://doi.org/10.1103/PhysRevX.11.021066